The daily newspaper “Prensa Austral” published out of Punta Arenas, Sub-Antarctic Chile, published an article in its Sunday science section entitled “The ancient plants of the Strait of Magellan: key pieces for investigating climate change”, referring to some of the inventories of the regional bryophyte and lichen flora we have contributed to. See: Prensa_Austral
The daily newspaper “Prensa Austral” published out of Punta Arenas, Sub-Antarctic Chile, published an article in its Sunday science section entitled “The ancient plants of the Strait of Magellan: key pieces for investigating climate change”, referring to some of the inventories of the regional bryophyte and lichen flora we have contributed to. See: Prensa_Austral
Author: Bernard Goffinet
Below your feet: new exhibit on mosses
 An exhibit of nine high resolution images of mosses from New England and Chile, printed on 5 ft metal sheets is on display at Wilbur Cross, on the UCONN campus. The material was collected and photographed by Bernard Goffinet and Mark Smith using the Macropod technology.
An exhibit of nine high resolution images of mosses from New England and Chile, printed on 5 ft metal sheets is on display at Wilbur Cross, on the UCONN campus. The material was collected and photographed by Bernard Goffinet and Mark Smith using the Macropod technology.
While the exhibit can be visited now, it will formally open after the constraints imposed by the pandemic have eased.
This exhibit was designed and installed by Collin Harty and funded by the Connecticut State Museum of Natural History, with additional support from a National Science Foundation grant entitled Collaborative research: Diversity of the moss Physcomitrium pyriforme: significance of autopolyploidy within a phylogenomic and experimental framework, awarded to Bernard Goffinet in the Department of Ecology & Evolutionary Biology (No. DEB-1753811).
Special thanks are extended to the Universidad de Magallanes, Instituto de Ecología & Biodiversidad, and Omora Ethnobotanical Park for facilitating collection of species in Chile.
New publication on lichens
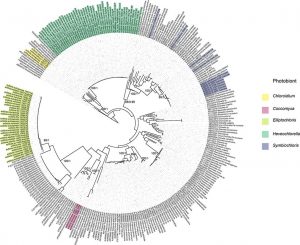 Complementing the study of the diversity of lichenized fungi of the Lobariaceae, is the study by Lindgren et al. exploring the patterns in the association in the two symbionts.
Complementing the study of the diversity of lichenized fungi of the Lobariaceae, is the study by Lindgren et al. exploring the patterns in the association in the two symbionts.
Lindgren H., B. Moncada, R. Lücking, N. Magain, A. Simon, B. Goffinet, E. Sérusiaux, M.P. Nelsen, J. Mercado-Díaz, T. Widhelm & T. Lumbsch. 2020. Cophylogenetic patterns in algal symbionts correlate with repeated symbiont switches during diversification and geographic expansion of lichen-forming fungi in the genus Sticta (Ascomycota, Peltigeraceae). Molecular Phylogenetics and Evolution 150: (in press). pdf Google scholar
Abstract reads: Species in the fungal genus Sticta form symbiotic associations primarily with either green algae or cyanobacteria, but tripartite associations or photosymbiodemes involving both types of photobionts occur in some species. Sticta is known to associate with green algae in the genus Symbiochloris. However, previous studies have shown that algae from other genera, such as Heveochlorella, may also be suitable partners for Sticta. We examined the diversity of green algal partners in the genus Sticta and assessed the patterns of association between the host fungus and its algal symbiont. We used multi-locus sequence data from multiple individuals collected in Australia, Cuba, Madagascar, Mauritius, New Zealand, Reunion and South America to infer phylogenies for fungal and algal partners and performed tests of congruence to assess coevolution between the partners. In addition, event-based methods were implemented to examine which cophylogenetic processes have led to the observed association patterns in Sticta and its green algal symbionts. Our results show that in addition to Symbiochloris, Sticta associates with green algae from the genera Chloroidium, Coccomyxa, Elliptochloris and Heveochlorella, the latter being the most common algal symbiont associated with Sticta in this study. Geography plays a strong role in shaping fungal-algal association patterns in Sticta as mycobionts associate with different algal lineages in different geographic locations. While fungal and algal phylogenies were mostly congruent, event-based methods did not find any evidence for cospeciation between the partners. Instead, the association patterns observed in Sticta and associated algae, were largely explained by other cophylogenetic events such as host-switches, losses of symbiont and failure of the symbiont to diverge with its host. Our results also show that tripartite associations with green algae evolved multiple times in Sticta.
The lab at Botany 2020
 The annual meeting of the Botanical Society of America starts tomorrow. Initially planned to take place in Alaska, it was converted into a virtual conference. The lab is contributing (to) four presentations:
The annual meeting of the Botanical Society of America starts tomorrow. Initially planned to take place in Alaska, it was converted into a virtual conference. The lab is contributing (to) four presentations:
Regular talk:
Patel, N., R. Medina, M.G. Johnson & B. Goffinet. 2020. Autopolyploidy contributes to cryptic speciation in mosses. Abstract 738.
Lightning talks:
Williams L.,N. Patel, R. Medina, B. Goffinet& M. Johnson. 2020. Methods to delimit speciation and determine population parameters of the moss Physcomitrium pyriforme using target capture sequencing. Abstract 400.
Buckowing, K., Y. Liu, A.J. Shaw, B.Goffinet, N.J. Wickett & M.G. Johnson. 2020. Expanded phylotranscriptomic sampling reveals gene family expansion in pleurocarpous mosses. Abstract 523.
Poster:
Anguloa J., M.G. Johnson, L. Pokorny, R. Medina, Y. Liu, B. Goffinet, A.J. Shaw & N.J. Wickett. 2020. Reconstructing the rapid radiation of pleurocarpous mosses using 802 nuclear genes. Abstract 605.
New publication on HGT from fungi to mosses
Sun G., S. Bai, Y. Guan, Q. Wang, S. Wang, Y. Liu, H. Liu, B. Goffinet, Y. Zhou, M. Paoletti, X. Hu, F. Haas, N. Fernendez-Pozo, A. Czyrt, H. Sun, S. Rensing & J. Huang. 2020. Are fungi-derived genomic regions related to antagonism toward fungi in mosses? New Phytologist 228: 1169–1175. pdf
In this article the authors “report two genomic regions in the nuclear genome of the moss Physcomitrium patens, previously Physcomitrella patens (Medina et al., 2019; Rensing et al., 2020), that contain mostly fungi-specific genes and mobile genetic elements. These two regions were identified in our genome screening for horizontally acquired genes in P. patens. Available evidence indicates that these fungi-specific genes are likely involved in the interaction between mosses and fungi. We discuss how these fungi-specific genes might have contributed to the defense against fungal and other microbial pathogens, as well as the loss of MFAs in mosses.”
On-line article on Sub-Antarctic bryophytes and lichens
 Introduction to article “Los pioneros vegetales del Estrecho de Magallanes” published on-line by the NGO Laderasur reads (google translation): Thousands of years ago, the Strait of Magellan was not what we know now, but rather an ice field. Thanks to the deglaciation, large lakes originated and the soils were covered with the pioneering organisms of the place: liquids, bryophytes and fungi. In this article, the scientists Laura Sánchez-Jardón, Bernard Goffinet, Ricardo Rozzi and the photographer Felipe Soza describe some representatives of these pioneers from the subantarctic region of Chile, relating them to the process of human colonization, recounting three successive discoveries that were made in the Strait of Magellan.
Introduction to article “Los pioneros vegetales del Estrecho de Magallanes” published on-line by the NGO Laderasur reads (google translation): Thousands of years ago, the Strait of Magellan was not what we know now, but rather an ice field. Thanks to the deglaciation, large lakes originated and the soils were covered with the pioneering organisms of the place: liquids, bryophytes and fungi. In this article, the scientists Laura Sánchez-Jardón, Bernard Goffinet, Ricardo Rozzi and the photographer Felipe Soza describe some representatives of these pioneers from the subantarctic region of Chile, relating them to the process of human colonization, recounting three successive discoveries that were made in the Strait of Magellan.
Visit article for nice pictures! Below is an English translation:
Thousands of years ago, the Strait of Magellan was not what we know now, but rather an ice field. Thanks to the deglaciation, large lakes originated and the soils were covered with the pioneering organisms of the place: liquids, bryophytes and fungi. In this article, scientists Laura Sánchez-Jardón, Bernard Goffinet, Ricardo Rozzi and photographer Felipe Soza describe some of the representatives of these pioneers from the region where they were made in the Strait of Magellan.
Twenty thousand years ago the Strait of Magellan was not such, but a vast field of ice. During the deglaciation started some sixteen thousand years ago, the retreat of the ice in its central part and surrounding areas gave rise to large lakes and terrestrial ecosystems whose rocks and soils were progressively covered by pioneering organisms: lichens, bryophytes and fungi.
This first discovery of the Strait of Magellan, associated with the melting of the ice cover, was producing unique processes of ecological succession led by small colonizing organisms, plant pioneers that generated the first ecosystems with a unique biota whose exuberance at the “end of the world” it is greater than in any other region of the planet.
When Hernando de Magallanes arrived at the southern tip of America five hundred years ago, there was a second discovery of the biota of the strait that today bears his name. The promise of riches offered by the New World, Terra Incognita, motivated scientists to explore the region, who were surprised by the abundance and diversity of lichens, mosses and fungi that grew on the ground and the trunks of subantarctic forests.
Some of these unique species are Ganoderma australe, which grows on coigües, lengas and ñires and is striking for its tongue or ear shape; in fact, they are known as “stick ears”; Cortinarius magellanicus, whose surprising color (bright purple) awakens the imaginations of scientists, tourists, and various Magellan co-inhabitants; some are even associated with algae and form lichens: Peltigera patagonica, one of the few species that appears on the tree line in the upper part of the mountain range and whose endemic distribution in the subantarctic region makes this species characteristic of it; Protousnea magellanica or “old man’s beard”, as this lichen is commonly called, abundant and widely distributed in subantarctic forests; along with Pseudocyphellaria berberina, they are endemic to the Southern Cone of South America.
In February 1834, Charles Darwin continued his navigation through the strait towards Puerto del Hambre aboard the Beagle, exploring the Shoal cove sector along the way. The British noted that this landscape represents a transition zone between aridity and humidity, between «Patagonia and Tierra del Fuego; many plants from these regions grow here ». The next day they arrived in Puerto del Hambre and on February 6 of that year, the young naturalist made his memorable ascent to Mount Tarn, recording in his Diary: I found a second species in other beech species in Chile; and Dr. Hooker informs me that a third species has recently been discovered in Van Diemen’s Land [Tasmania]. How unique is this relationship between parasitic fungi and the trees on which they grow, in the farthest parts of the world! In Tierra del Fuego, the fungus in its smooth and mature state is collected in large quantities by women and children, and is eaten without cooking.
The young naturalist was referring here to the digueñes or dihueñes (Cyttaria spp.), Also recognized in other scientific expeditions by the southern hemisphere during the 19th century. These endemic fungi, metaphorically called “Indian bread”, that grew on the trunks and branches of the coigüe and lenga, both trees of the dominant Nothofagus genus in the region, were already discovered by the first human populations that reached the strait and other areas from the Magellan subantarctic ecoregion, who found in them a unique food source.
Indeed, little by little, the extreme richness of fungi was revealed, as well as the exceptional diversity of small plants in the sub-Antarctic region of the Strait of Magellan: mosses, liverworts and hornworts, which together are called bryophytes. These small organisms facilitated the recolonization of life once the ice was removed in the Strait of Magellan, sheltering in the neighboring territories the original peoples that arrived more than ten thousand years ago.
Just twenty years ago, a third discovery of the Magallanes region south of the strait, in the Cape Horn archipelagos, happened fortuitously, leading to the identification of this sub-Antarctic region as a world center of diversity of bryophytes and lichens. The unique biodiversity that inhabits these archipelagos is of great value for life on the planet and we must jointly take care of this biota and ecosystems that contribute to planetary health.
Among these species are the Dendroligotrichum dendroides or “pinito moss”, with the appearance of a miniature tree – up to 20 centimeters the largest -; Bartramia mossmaniana; Sphagnum magellanicum, an endemic species of the southern hemisphere that proliferates locally on accumulations of dead organic matter, forming a particular type of ecosystem called «peatland», extraordinarily important in carbon fixation and in the retention of water and nutrients; in especially humid and shady places are also found hornwort like Phaeomegaceros chiloensis.
These miniature forests of fungi, lichens and bryophytes, in addition to all the associated fauna of invertebrates, have accompanied the three discoveries of the Strait of Magellan. From an ethical point of view, they can be considered as Magellanic co-inhabitants: literally, they have shared the habitats of the strait with humans since the first settlement and have had a leading role in their food, health, fire source, water, culture and , lately, in the scientific vanguard. From the south of the world, the knowledge of the great Magellanic flora will contribute to its conservation and the sustainability of the planet in the scenario of global socio-environmental change. About the authors: Laura Sanchez-Jardón, is a Doctor in ecology and environment at the Complutense University of Madrid; Ricardo Rozzi is a Doctor in Ecology and a Master in Philosophy from the University of Connecticut; Bernard Goffinet is a Doctor in Botany University of Connecticut and Jorge Felipe Soza Soza (Instagram: @felipesozaphotography)
New publication on comparative transcriptomics
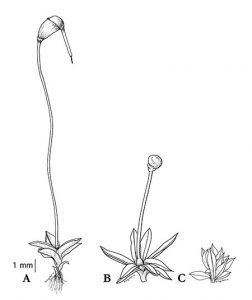
Kirbis A., M. Waller, M. Ricca, Z. Bont, A. Neubauer, B. Goffinet & P. Szövényi. 2020. The transcriptomic landscape of differential sporophyte development in two mosses, Physcomitrium (Physcomitrella) patens and Funaria hygrometrica. Frontiers in Plant Sciences11: 747. pdf
Abstract reads: Understanding the molecular basis of morphological shifts is a fundamental question of evolutionary biology. New morphologies may arise through the birth/death of genes (gene gain/loss) or by reutilizing existing gene sets. Yet, the relative contribution of these two processes to radical morphological shifts is still poorly understood. Here, we use the model system of two mosses, Funaria hygrometrica and Physcomitrium (Physcomitrella) patens, to investigate the molecular mechanisms underlying contrasting sporophyte architectures. We used comparative analysis of time-series expression data for four stages of sporophyte development in both species to address this question in detail. We found that large-scale differences in sporophytic architecture are mainly governed by orthologous (i.e., shared) genes frequently experiencing temporal gene expression shifts between the two species. While the absolute number of species-specific genes expressed during sporophyte development is somewhat smaller, we observed a significant increase of their proportion in preferentially sporophyte expressed genes, suggesting a fundamental role in the sporophyte phase. However, further functional studies are necessary to determine their contribution to diverging sporophyte morphologies. Our results add to the growing set of studies suggesting that radical changes in morphology may rely on the heterochronic expression of conserved regulators.
Two new publications on lichens

We propose a new genus to accommodate some species of lichenized fungi from the New World, including the Southeastern United States, in a new genus, as part of the ongoing studies of the evolution of the Lobariaceae.
Simon A., R. Lücking, B. Moncada, J.A. Mercado-Díaz, F. Bungartz, M. Cáceres, E. Gumboski, S. Maria de Azevedo Martinsi, D. Parker & B. Goffinet. 2020. Emmanuelia, a new genus of lobarioid lichen-forming fungi (lichenized Ascomycota: Peltigerales). Plant and Fungal Systematics 65: 76–94. pdf
Abstract reads: The former family Lobariaceae, now included in Peltigeraceae as subfamily Lobarioideae, has undergone substantial changes in its generic classification in recent years, based on phylogenetic inferences highlighting the polyphyly of the speciose genera Lobaria, Pseudocyphellaria and Sticta. Here we introduce the new genus Emmanuelia, named in honor of Prof. Emmanuël Sérusiaux for his extensive work on the Peltigerales. Emmanuelia currently comprises twelve species. It is superficially similar to the lobarioid genus Ricasolia, but differs by its apothecia, rimmed by overarching and often crenulate to lobulate margins, with the parathecium (proper excipulum) and the amphithecium (thalline excipulum formed by the thallus cortex) apically separated and of a different structure. Also, ascospore dimensions and shape differ between the two genera, with the ascospores of Emmanuelia being longer and narrower. Molecular phylogenetic analyses using DNA nucleotide sequences of the internal transcribed spacer region (ITS) and the small subunit of mitochondrial ribosomal DNA (mtSSU) confirm that Emmanuelia belongs to the Lobaria s.lat. clade and forms a monophyletic group sister to the lineage consisting of Dendriscosticta, Lobariella and Yoshimuriella. None of the available generic names of lobarioid lichens can be applied to this group, and consequently a new name is proposed for this new genus, which is typified with E. ravenelii comb. nov. Eleven other species are transferred to Emmanuelia: E. americana comb. nov., E. conformis comb. nov., E. cuprea comb. nov., E. elaeodes comb. nov., E. erosa comb. nov., E. excisa comb. nov., E. lobulifera comb. nov., E. ornata comb. nov., E. patinifera comb. nov., E. pseudolivacea comb. nov. and E. tenuis comb. nov. The genus is represented in North America by three species, including E. lobulifera, which is resurrected from synonymy with E. (Lobaria) tenuis, a South American species, and E. ornata, whose populations were previously treated under E. (Lobaria) ravenelii.
Suspected to represent a new species, populations of Peltigera from Papua New Guinea are now recognized as P. serusiauxii: Magain N., B. Goffinet, A. Simon, J. Seelan Sathiya, I. Medeiros, F. Lutzoni & J. Miadlikowska. 2020. Peltigera serusiauxii, a new species in section Polydactylon from Papua New Guinea and Malaysia (Lecanoromycetes, Ascomycota). Plant and Fungal Systematics 65: 139–146. pdf
Abstract reads: Peltigera serusiauxii is proposed here as a new species from Papua New Guinea and Sabah, northern Borneo (Malaysia). The species belongs to the polydactyloid clade of section Polydactylon. Because of its large thalli with a glabrous upper surface, this species was previously identified as P. dolichorhiza, but it differs by its polydactylon-type lower surface and the high amount of dolichorrhizin. It appears to be a strict specialist in its association with Nostoc phylogroup IX throughout its known distribution. This is one of many undescribed species remaining to be formally described within the genus Peltigera,especially in Asia and Australasia.
New publication on tardigrades!
It started with a search for bryophyte fragments in the feces of high Andean birds on Navarino island (see post) when Michael Robertson and Nicholas Russo (former EEB student now at UCLA) discovered tardigrades in the samples. Their observation are now published in Polar Biology.
Robertson M.W., N.J. Russo, S.J. McInnes, B. Goffinet & J.E. Jiménez. 2020. Potential dispersal of tardigrades by birds through endozoochory: evidence from sub-Antarctic White-bellied Seedsnipe (Attagis malouinus). Polar Biology in press.
Abstract reads: Tardigrades are potentially dispersed by birds, but the extent of the interactions between birds and tardigrades is virtually unknown. We discovered nine tardigrades within feces of White-bellied Seedsnipe (Attagis malouinus) collected from high Andean tundra on Navarino Island, Chile. Eight of the tardigrade specimens began moving once rehydrated. Two specimens belonged to the genus Adropion (Hypsibiidae), one to the Macrobiotus (Macrobiotidae), and five could not be identified. A ninth specimen was a species of Isohypsibius in an embryonic egg state. These tardigrades could have passed through the avian digestive tract after incidental ingestion or burrowed into the feces post-defecation to feed on microorganisms and undigested plant matter present in the feces. To our knowledge, this is the first discovery of tardigrades in bird feces and may have implications for tardigrade distributions if birds transport tardigrades endogenously.
New publication on bryophytes
Juan Carlos Villarreal published the last chapter of his dissertation, focused on the population genetic study of Nothoceros aenigmaticus, a clonal hornwort with allopatric sexual populations. The study was picked up and completed by Juan Carlos’ postdoc, Marta Alonso Garcia, who had visited our lab when she was finishing her Ph.D. in Murcia, Spain. Alonso Garcia M., Villarreal J.C., K. McFarland & B. Goffinet. 2020. Population genomics confirms extreme sex ratio of a clonal bryophyte. Frontiers in Plant Science 11: 495. pdf
Abstract reads: The southern Appalachian (SA) is one of the most biodiversity−rich areas in North America and has been considered a refugium for many disjunct plant species, from the last glacial period to the present. Our study focuses on the SA clonal hornwort, Nothoceros aenigmaticus J.C. Villarreal & K.D. McFarland. This hornwort was described from North Carolina and is widespread in the SA, growing on rocks near or submerged in streams in six and one watersheds of the Tennessee (TR) and Alabama (AR) Rivers, respectively. Males and female populations occur in different watersheds, except in the Little Tennessee (TN) River where an isolated male population exists ca. 48 km upstream from the female populations. The sex ratio of 1:0 seems extreme in each population. In this study, we use nuclear and organellar microsatellites from 250 individuals from six watersheds (seven populations) in the SA region and two populations from Mexico (23 individuals). We, then, selected 86 individuals from seven populations and used genotyping by sequencing to sample over 600 bi-allelic markers. Our results suggest that the SA N. aenigmaticus and Mexican plants are a nested within a clade of sexual tropical populations. In the US populations, we confirm an extreme sex ratio and only contiguous US watersheds share genotypes. The phylogenetic analysis of SNP data resolves four clusters: Mexican populations, male plants (Little Pigeon and Pigeon river watersheds) and two clusters of female plants; one from the Little Tennessee and Hiwassee Rivers (TR) and the other from the Ocoee (TR) and Coosa (AR) Rivers. All clusters are highly differentiated (Fst values over 0.9). In addition, our individual assignment analyses and PCAs reflect the phylogenetic results grouping the SA samples in three clades and recovering males and female plants with high genetic differentiation (Fst values between 0.5 and 0.9 using microsatellites and bi-allelic markers). Our results point to Pleistocene events shaping the biogeographical pattern seen in US populations. The extreme sex ratio reflects isolation and highlights the high vulnerability of the populations in the SA.