Hu R., X. Li, Y. Hu, R. Zhang, Q. Lv, M. Zhang, X. Sheng, F. Zhao, Z. Chen, Y. Ding, H. Yuan, X. Wu, S. Xing, X. Yan, F. Bao, P. Wan, L. Xiao, X. Wang, W. Xiao, E.L. Decker, N. van Gessel, H. Renault, G. Wiedemann, N.A. Horst, F.B. Haas, P.K.I. Wilhelmsson, K. Ullrich, E. Neumann, B. Lv, C. Liang, H. Du, H. Lu, Q. Gao, Z. Cheng, H. You, P. Xin, J. Chu, C.-H. Huang, Y. Liu, S. Dong, L. Zhang, F. Chen, L. Deng, F. Duan, W. Zhao, K. Li, Z. Li, X. Li, H. Cui, Y. Zhang, C. Ma, R. Zhu, Y. Jia, M. Wang, M. Hasebe, J. Fu, B. Goffinet, H. Ma, S.A. Rensing, R. Reski, Y. He. 2023. Adaptive evolution of the enigmatic Takakia now facing climate change in Tibet. Cell 186: 3558–3576. pdf
Hu R., X. Li, Y. Hu, R. Zhang, Q. Lv, M. Zhang, X. Sheng, F. Zhao, Z. Chen, Y. Ding, H. Yuan, X. Wu, S. Xing, X. Yan, F. Bao, P. Wan, L. Xiao, X. Wang, W. Xiao, E.L. Decker, N. van Gessel, H. Renault, G. Wiedemann, N.A. Horst, F.B. Haas, P.K.I. Wilhelmsson, K. Ullrich, E. Neumann, B. Lv, C. Liang, H. Du, H. Lu, Q. Gao, Z. Cheng, H. You, P. Xin, J. Chu, C.-H. Huang, Y. Liu, S. Dong, L. Zhang, F. Chen, L. Deng, F. Duan, W. Zhao, K. Li, Z. Li, X. Li, H. Cui, Y. Zhang, C. Ma, R. Zhu, Y. Jia, M. Wang, M. Hasebe, J. Fu, B. Goffinet, H. Ma, S.A. Rensing, R. Reski, Y. He. 2023. Adaptive evolution of the enigmatic Takakia now facing climate change in Tibet. Cell 186: 3558–3576. pdf
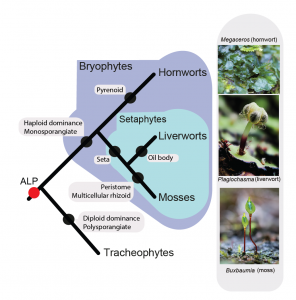
The most extreme environments are the most vulnerable to transformation under a rapidly changing climate. These ecosystems harbor some of the most specialized species, which will likely suffer the highest extinction rates. We document the steepest temperature increase (2010–2021) on record at altitudes of above 4,000 m, triggering a decline of the relictual and highly adapted moss Takakia lepidozioides. Its de-novo-sequenced genome with 27,467 protein-coding genes includes distinct adaptations to abiotic stresses and comprises the largest number of fast-evolving genes under positive selection. The uplift of the study site in the last 65 million years has resulted in life-threatening UV-B radiation and drastically reduced temperatures, and we detected several of the molecular adaptations of Takakia to these environmental changes. Surprisingly, specific morphological features likely occurred earlier than 165 mya in much warmer environments. Following nearly 400 million years of evolution and resilience, this species is now facing extinction.
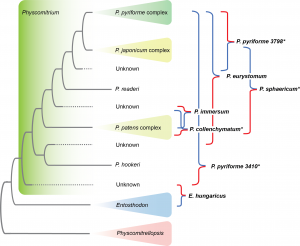
 Zhang L., Q. Zuo, W.Z. Ma, J. Shevock, N. Patel, M. Johnson, R. Medina, N. Wilding & B. Goffinet. 2023. Phylogenomics resolves the rediscovered Himalayan endemic Brachymeniopsis gymnostoma (Bryophyta, Funariaceae), as a species of Entosthodon, prompting also the transfer of Clavitheca poeltii. Taxon 72: 1216–1227. pdf Google Scholar
Zhang L., Q. Zuo, W.Z. Ma, J. Shevock, N. Patel, M. Johnson, R. Medina, N. Wilding & B. Goffinet. 2023. Phylogenomics resolves the rediscovered Himalayan endemic Brachymeniopsis gymnostoma (Bryophyta, Funariaceae), as a species of Entosthodon, prompting also the transfer of Clavitheca poeltii. Taxon 72: 1216–1227. pdf Google Scholar Zhang L., Q. Zuo, W.Z. Ma, J. Shevock, N. Patel, M. Johnson, R. Medina, N. Wilding & B. Goffinet. 2023. Phylogenomics resolves the rediscovered Himalayan endemic Brachymeniopsis gymnostoma (Bryophyta, Funariaceae), as a species of Entosthodon, prompting also the transfer of Clavitheca poeltii. Taxon 72: 1216–1227. pdf Google Scholar
Zhang L., Q. Zuo, W.Z. Ma, J. Shevock, N. Patel, M. Johnson, R. Medina, N. Wilding & B. Goffinet. 2023. Phylogenomics resolves the rediscovered Himalayan endemic Brachymeniopsis gymnostoma (Bryophyta, Funariaceae), as a species of Entosthodon, prompting also the transfer of Clavitheca poeltii. Taxon 72: 1216–1227. pdf Google Scholar