 Dong S., J.-Y. Xue, S. Zhang, L. Zhang, H. Wu, Z. Chen, B. Goffinet & Yang Liu. 2018. Complete mitochondrial genome sequence of Anthoceros angustus: conservative evolution of mitogenome in hornworts. The Bryologist 121: 14–22. pdf
Dong S., J.-Y. Xue, S. Zhang, L. Zhang, H. Wu, Z. Chen, B. Goffinet & Yang Liu. 2018. Complete mitochondrial genome sequence of Anthoceros angustus: conservative evolution of mitogenome in hornworts. The Bryologist 121: 14–22. pdf
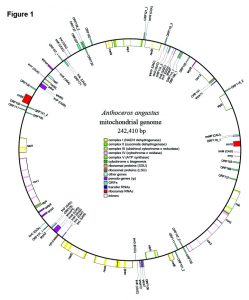
Abstract reads: The architecture and composition of the mitochondrial genome of bryophytes is currently considered much more stable and invariant than that of vascular plants. For example, liverworts and mosses appear to have nearly static mitochondrial genomes, whereas neither of two vascular plant mitogenomes sequenced to date share the same gene order. By contrast, the two available mitogenomes of hornworts, Nothoceros aenigmaticus and Phaeoceros laevis, show high consistency in gene content but differ in their gene order. To explore the patterns of hornwort mitogenome structure at a broader phylogenetic breadth and provide insights into plant mitogenome evolution, we assembled the complete mitochondrial genome from a third hornwort species, Anthoceros angustus Steph. Its mitogenome comprises 242,410 base pairs, and encodes 21 protein coding genes, three rRNA genes and 20 tRNA genes, with 38 introns disrupting 16 protein-coding genes, including three newly described introns, making it the most intron-rich plant mitochondrial genome determined to date. A total of ten putatively pseudogenized protein coding genes were found in Anthoceros; eight of them are shared with Nothoceros and Phaeoceros. The genic composition of the Anthoceros mitogenome differs from that of the other two hornworts by only one or two gene insertions or losses. The gene order in the Anthoceros mtDNA differs from that in Phaeoceros and Nothoceros by only one and three events of inversion or translocations, respectively. Overall, hornworts appear to have kept a stable mt genome structure, which could have characterized early land plant mt genome evolution.